XB-FEAT-953844: Difference between revisions
| Line 9: | Line 9: | ||
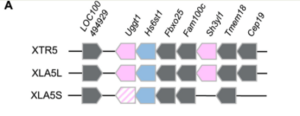
''uggt1.L and sh3yl1.L'', show expression levels similar to ''hs6st1.L'' (see Figs. S5B and D). Because these two genes are located nearer the ''hs6st1.L'' locus than to ''cep19.L'', it is suggested that ''hs6st1.L, uggt1.L,'' and ''sh3yl1.L'', share cis-regulatory modules, and that these genomic regions were lost or changed during Xla.S chromosome evolution, leading to a change in ''hs6st1.S'' expression pattern." | ''uggt1.L and sh3yl1.L'', show expression levels similar to ''hs6st1.L'' (see Figs. S5B and D). Because these two genes are located nearer the ''hs6st1.L'' locus than to ''cep19.L'', it is suggested that ''hs6st1.L, uggt1.L,'' and ''sh3yl1.L'', share cis-regulatory modules, and that these genomic regions were lost or changed during Xla.S chromosome evolution, leading to a change in ''hs6st1.S'' expression pattern." | ||
Synteny FigureS5A from Yamamoto et al 2023 | |||
[[File:Screenshot 2024-05-14 at 11.36.25 AM.png|thumb]] | |||
Latest revision as of 08:40, 14 May 2024
hs6st1
This is the community wiki page for the gene hs6st1 please feel free to add any information that is relevant to this gene that is not already captured elsewhere in Xenbase
notes on synteny and gene evolution/function
source: Yamamoto et al 2023 [1]
synteny patterns " around Hs6st1 genes are well conserved around the [Xla.hs6st1.L] gene and the X. tropicalis hs6st1 gene. However, three [S homeolog] genes, uggt1.S, sh3yl1.S, cep19.S, were lost from the Xla.S chromosome (see Fig. S5A).
uggt1.L and sh3yl1.L, show expression levels similar to hs6st1.L (see Figs. S5B and D). Because these two genes are located nearer the hs6st1.L locus than to cep19.L, it is suggested that hs6st1.L, uggt1.L, and sh3yl1.L, share cis-regulatory modules, and that these genomic regions were lost or changed during Xla.S chromosome evolution, leading to a change in hs6st1.S expression pattern."
Synteny FigureS5A from Yamamoto et al 2023